You signed in with another tab or window. Reload to refresh your session.You signed out in another tab or window. Reload to refresh your session.You switched accounts on another tab or window. Reload to refresh your session.Dismiss alert
I do not actually know WGD in detail, but I am in a situation where I have to do WGD analysis. So I want to use your pipeline.
Fortunately, this pipeline is easy to use.
But I have a problem that I can not solve.
Can you tell me how to check the WGD date after using this pipeline?
Many thanks for making very easy to understand script using Dup_gene finder software to find the WGD events. Please guide me about this command...print "Usage: DupGen_finder.pl -i data_directory -t target_species -c outgroup_species -o output_directory\n";
This command is about to attach the files to compare/find the wgd events types. I would like to find wgd types in five species. how I can re-write this command? Could you please explain to me. Thanks!
The DupGen_finder was developed to identify different modes of duplicated gene pairs in a given species. A detailed tutorial for DupGen_finder has been provided, please find more details here.
Herein, I briefly describe the usage. We used Arabidopsis thaliana (Ath, target_species) as an example to detect different modes of gene duplications. Firstly, you need to select an proper outgroup species for the target species (Arabidopsis). Here we used Nelumbo nucifera (Nnu, outgroup_species) as outgroup for Arabidopsis. Before running DupGen_finder, you need to prepare four pre-computed input files including Ath.gff, Ath.blast, Ath_Nnu.gff, and Ath_Nnu.blast, and put these files into the same folder (data_directory). And then you can run the following command to identify duplicated gene pairs:
DupGen_finder.pl -i data -t Ath -c Nnu -o results
The output files will be stored under this folder results.
If you want to detect the WGD events of different ages, please refer to the below tutorial:
Many thanks for your reply.
I am following your this script. So, I should follow all the steps starts from below...to end. Or need some changes? Please guide as I am, not good in Linux. Thanks!
You need not to change the code in the script DupGen_finder.pl. What you should do is to install DupGen_finder on your server (Linux system) and execute the script DupGen_finder.pl with input files on the server. Please find more details in the tutorial.
Please execute the below command to get help information about DupGen_finder:
Heartiest greetings and very happy National days to you. Now I have all the duplicated gene (types) and thanks to you of course. But now is the problem of how to make the figure from four species and five types of duplicated genes. Like Mcscan has the command to draw figure. Can you please help to draw figure or tell any commands after running Dup_gen_Finder. Thanks again!
Dear Qiao Xin:
Could you give some advise about how to chose a outgroup? Thanks.
Sorry for my delayed reply. The choice of outgroup species only have impact on the identification of transposed gene pairs. If you choose a closely related species as outgroup, less transposed-pairs will be identified. If you choose a distantly related species as outgroup, more transposed-pairs will be identified.
You can choose an appropriate outgroup according to your research project. For example, assuming that you are going to detect different modes of gene duplications in Arabidopsis thaliana,
If you choose Arabidopsis lyrata (a closely related species for A. thaliana) as outgroup (Fig. 1), the transposed duplications that occurred after A. thaliana-A. lyrata divergence will be identified.
If you choose Brassica rapa (a more distant species for A. thaliana) as outgroup, the transposed duplications that occurred after Arabidopsis-Brassica divergence will be identified.
If you choose Carica papaya (a distantly related species for A. thaliana) as outgroup, the transposed duplications that occurred after Brassicaceae-Carica divergence will be identified.
...
Therefore, different outgroup species can be used to detect transposed gene duplications that occurred within different epochs.
Heartiest greetings and very happy National days to you. Now I have all the duplicated gene (types) and thanks to you of course. But now is the problem of how to make the figure from four species and five types of duplicated genes. Like Mcscan has the command to draw figure. Can you please help to draw figure or tell any commands after running Dup_gen_Finder. Thanks again!
Sorry for my delayed reply. Matplotlib or pandas can be used to plot the stacked bar chart. The detailed examples and related code are as follows, and hope it works.
whether multiple species were selected as the outgroups used for analysis ?
Only one outgroup species is required for DupGen_finder to identify duplicated gene pairs in a given target/query sepecies. Sure, different outgroup species that have diverse genetic distance with the target species can be used to detect transposed gene duplications that occurred within different epochs.
Activity
qiao-xin commentedon Jul 1, 2019
I guess you have obtained the WGD-derived gene pairs, and you can estimate the WGD date by following the two steps below:
-- pipeline
-- pipeline
Gill85 commentedon Sep 26, 2019
Dear Qiao Xin,
Many thanks for making very easy to understand script using Dup_gene finder software to find the WGD events. Please guide me about this command...print "Usage: DupGen_finder.pl -i data_directory -t target_species -c outgroup_species -o output_directory\n";
This command is about to attach the files to compare/find the wgd events types. I would like to find wgd types in five species. how I can re-write this command? Could you please explain to me. Thanks!
qiao-xin commentedon Sep 26, 2019
Hi Rafaqat Ali Gill,
The DupGen_finder was developed to identify different modes of duplicated gene pairs in a given species. A detailed tutorial for DupGen_finder has been provided, please find more details here.
Herein, I briefly describe the usage. We used Arabidopsis thaliana (Ath, target_species) as an example to detect different modes of gene duplications. Firstly, you need to select an proper outgroup species for the target species (Arabidopsis). Here we used Nelumbo nucifera (Nnu, outgroup_species) as outgroup for Arabidopsis. Before running DupGen_finder, you need to prepare four pre-computed input files including Ath.gff, Ath.blast, Ath_Nnu.gff, and Ath_Nnu.blast, and put these files into the same folder (data_directory). And then you can run the following command to identify duplicated gene pairs:
The output files will be stored under this folder results.
If you want to detect the WGD events of different ages, please refer to the below tutorial:
-- A pipeline used to identify Ks peaks by fitting Gaussian Mixture Model (GMM)
Gill85 commentedon Sep 27, 2019
Dear Qiao Xin,
Many thanks for your reply.
I am following your this script. So, I should follow all the steps starts from below...to end. Or need some changes? Please guide as I am, not good in Linux. Thanks!
use Getopt::Std;
%options=();
getopts("i:t:o:c:d:k:g:s:e:m:w:a:x:", %options);
if(!exists $options{i} || !exists $options{t} || !exists $options{o} || !exists $options{c})
{
print "Usage: DupGen_finder.pl -i Genome_duplication_practice -t Ath -c Nnu -o results\n";
qiao-xin commentedon Sep 27, 2019
Hi Rafaqat Ali Gill,
You need not to change the code in the script DupGen_finder.pl. What you should do is to install DupGen_finder on your server (Linux system) and execute the script DupGen_finder.pl with input files on the server. Please find more details in the tutorial.
Please execute the below command to get help information about DupGen_finder:
or
Good luck!
Gill85 commentedon Sep 27, 2019
Dear Qiao Xin,
Many thanks
Gill85 commentedon Oct 2, 2019
Dear Qiao Xin,
Heartiest greetings and very happy National days to you. Now I have all the duplicated gene (types) and thanks to you of course. But now is the problem of how to make the figure from four species and five types of duplicated genes. Like Mcscan has the command to draw figure. Can you please help to draw figure or tell any commands after running Dup_gen_Finder. Thanks again!
ncgrjsmiao commentedon Dec 5, 2019
Dear Qiao Xin:
Could you give some advise about how to chose a outgroup? Thanks.
qiao-xin commentedon Apr 16, 2020
Sorry for my delayed reply. The choice of outgroup species only have impact on the identification of transposed gene pairs. If you choose a closely related species as outgroup, less transposed-pairs will be identified. If you choose a distantly related species as outgroup, more transposed-pairs will be identified.
You can choose an appropriate outgroup according to your research project. For example, assuming that you are going to detect different modes of gene duplications in Arabidopsis thaliana,
If you choose Arabidopsis lyrata (a closely related species for A. thaliana) as outgroup (Fig. 1), the transposed duplications that occurred after A. thaliana-A. lyrata divergence will be identified.
If you choose Brassica rapa (a more distant species for A. thaliana) as outgroup, the transposed duplications that occurred after Arabidopsis-Brassica divergence will be identified.
If you choose Carica papaya (a distantly related species for A. thaliana) as outgroup, the transposed duplications that occurred after Brassicaceae-Carica divergence will be identified.
...
Therefore, different outgroup species can be used to detect transposed gene duplications that occurred within different epochs.
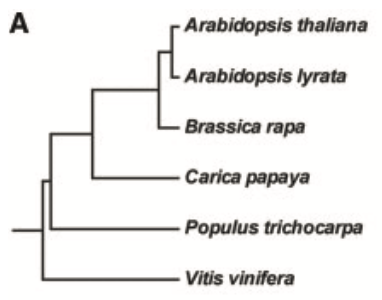
Fig. 1qiao-xin commentedon Apr 16, 2020
Sorry for my delayed reply. Matplotlib or pandas can be used to plot the stacked bar chart. The detailed examples and related code are as follows, and hope it works.
Matplotlib, example 1
pandas, example 2
Sh1ne111 commentedon Jul 6, 2021
whether multiple species were selected as the outgroups used for analysis ?
qiao-xin commentedon Jul 7, 2021
Only one outgroup species is required for DupGen_finder to identify duplicated gene pairs in a given target/query sepecies. Sure, different outgroup species that have diverse genetic distance with the target species can be used to detect transposed gene duplications that occurred within different epochs.