-
Notifications
You must be signed in to change notification settings - Fork 119
How to map cell fate to branches? #219
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Comments
|
Hi @ClarkGregg sorry for the delay in response. I will answer your questions in the following: |
|
Hello, Following your explanation of BEAM
So pre-branch in case of testing node 3 should rather be the state 1 branch as it is leading towards pseudotime 0 ? |
|
Hi@Xiaojieqiu. Sincere thanks for your explanation. May I ask why the pre-branch will only include cells from state 2 when testing for the difference between state4 and state7. Thanks!
|
|
I have same question. Why only state 2 is the pre-branch? |




Dear all.
I am working on single cell RNA-seq data using your R package: Monocle. While I am performing pseudotime analysis, I have some questions, hopefully you can clear my mind.
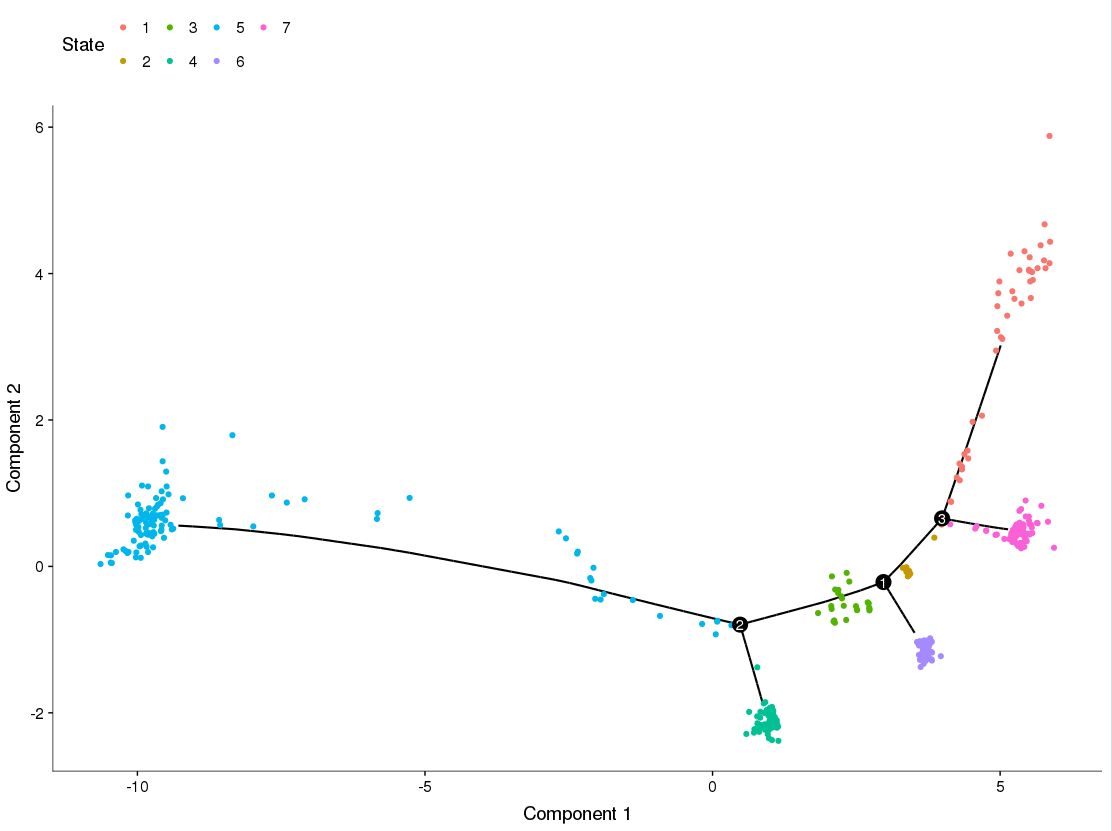
By following the pipeline posted on the website, I constructed a trajectory with several branches (States). Then I sought to perform differential expression test to identify genes expressed in a branch-dependent manner. Here are my question one: I used BEAM() function inside Monocle to do it, I tried to compare state 1 and state 7, or equally branch point 3 and visualize my result using plot_genes_branched_heatmap() function. I am wondering, if the 'pre-branch' state is comprised of all states excepted for the two being tested or just state 2, in this particular case. And also what is the 'pre-branch' if I try to compare states that sit within different branches, say, branch 4 and branch 7.
My question 2 is about a parameter, the parameter inside plot_genes_branched_heatmap() called branch_label. the default branch label is 'cell fate 1' and 'cell fate 2'. How do I map these labels to the corresponding branches being tested? For example, if I am comparing branch 1 and branch 7. should 'cell fate 1' correspond to branch 1 and branch 7 correspond to 'cell fate 2' ?
The attachment is the trajectory I constructed.
Thank you in advance
The text was updated successfully, but these errors were encountered: