```r library(TCGAbiolinks) query_SNV <- GDCquery(project = "TCGA-LIHC", data.category = "Simple Nucleotide Variation", data.type = "Masked Somatic Mutation", workflow.type = "Aliquot Ensemble Somatic Variant Merging and Masking") GDCdownload(query_SNV) mafFilePath2 = dir(path = "GDCdata/TCGA-LIHC",pattern = "masked.maf.gz$",full.names = T,recursive=T) library(maftools) mafdata2 <- lapply(mafFilePath2, function(x) {read.maf(x,isTCGA = T)}) snv_data2 = merge_mafs(mafdata2) ``` 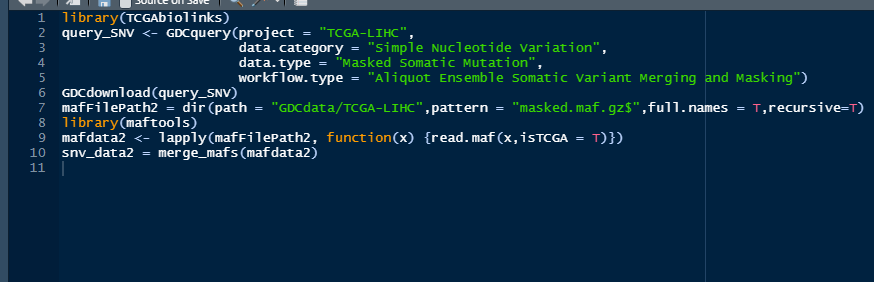  Why this error occurred, thank you for your answer.
Activity
PoisonAlien commentedon Jun 4, 2022
Hi,
I couldn't figure out the issue. But try the below workaround..
Hope this helps.
li910493462 commentedon Sep 20, 2022
i have the same error,please help me,thank you
github-actions commentedon Aug 18, 2023
This issue is stale because it has been open for 60 days with no activity.
github-actions commentedon Sep 1, 2023
This issue was closed because it has been inactive for 14 days since being marked as stale.