-
Notifications
You must be signed in to change notification settings - Fork 149
Closed
Labels
documentationImprovements or additions to documentationImprovements or additions to documentation
Description
hi!
When i run filterDoublets(ArchRProj = proj1),I got an error "Error in as.POSIXct.numeric(e) : 'origin' must be supplied".
And i don't konw how to solve the problem.There is nothing wrong with the previous steps.
I want to know how to deal with this problem, continue to analyze the data (My data comes from 10x)
Thanks!
Metadata
Metadata
Assignees
Labels
documentationImprovements or additions to documentationImprovements or additions to documentation
Type
Projects
Milestone
Relationships
Development
Select code repository
Activity
rcorces commentedon Jun 19, 2020
Can you reproduce this error using the tutorial dataset?
xuzongren commentedon Jun 22, 2020
When i use tutorial dataset,this error does not occur.
Is there something wrong with my data? (but my data comes from 10X cellranger-atac,I didn't do anything else to fragments.tsv.gz file.)
How can I deal with this error?
Meggie-MH commentedon Jun 22, 2020
Run into the same error here. Hope to see update on the issue.
rcorces commentedon Jun 22, 2020
We will need more information in order to help since we cannot reproduce this error. If you can create a minimal example, including input files, that would be helpful.
xuzongren commentedon Jun 23, 2020
The R^2 values of my samples all < 0.9, that mean I can skip this step(filterDoublets)?
rcorces commentedon Jun 23, 2020
No, that is not the correct interpretation. see #173 (comment) for an explanation
xuzongren commentedon Jun 23, 2020
Thanks. I've read the question( #173 (comment)).
And how can I create a minimal example so that you can better solve this problem?
rcorces commentedon Jun 23, 2020
Ideally, take a random 50,000 lines (or the first 50,000 if you prefer) of your fragment file and see if you get the same error. If so, provide us with the small fragment file and the code you used that resulted in the error.
jgranja24 commentedon Jun 23, 2020
When does this happen? Towards the end or beginning? Is there a logfile you can share?
xuzongren commentedon Jun 23, 2020
@jgranja24 ,This step does not generate any logfiles.
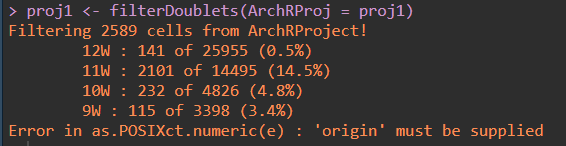
it show me this:
xuzongren commentedon Jun 24, 2020
@rcorces ,I'm sorry for what I said earlier(I guess I forgot to run the tutorial dataset)
Today,I run tutorial dataset, the same error occurred……emmmm,do you have any suggestions?
Thanks!
rcorces commentedon Jun 24, 2020
@Meggie-MH are you also running on Windows? We arent currently supporting windows and I should make that more clear on the home page.
@jgranja24 - I think this is a problem related to date format conversion on Windows from Sys.time(). https://rdrr.io/r/base/Sys.time.html
ArchR/R/FilterCells.R
Line 101 in efc743c
Meggie-MH commentedon Jun 24, 2020
@rcorces No, I was running on Mac. I also tried the tutorial dataset and ran into the same error from filterDoublets. The only thing I did different from the tutorial was creating arrow files by inputting individual fragments.tsv.gz and then combined all the arrow files into one character vector before running the following the steps. For some reasons, it takes infinite time to createarrowfiles directly from 'hemefragments' folder which contains three tsv.gz datasets. Not sure if that's the step starts with this issue.
9 remaining items
Meggie-MH commentedon Jun 25, 2020
@rcorces Woohoo, rewrite the function works! Thanks a lot :)
huidongchen commentedon Jun 25, 2020
First of all, thanks for creating such an amazing tool! I have been such a huge fan.
Unfortunately I'm running into the same problem. The same error happens when filtering doublets. (I m running it on Linux)
proj <- filterDoublets(ArchRProj = proj,cutEnrich=5)Error info
Rewriting the function as suggested by @jgranja24 works for me too.
rcorces commentedon Jun 25, 2020
@Meggie-MH - thanks for checking. We will update the dev branch with the fix from @jgranja24
@huidongchen - can you confirm which version of R you are using? Presumably >4.0?
huidongchen commentedon Jun 25, 2020
Thanks for your quick reply! Yes, I'm using R 4.0.1
xuzongren commentedon Jun 28, 2020
@jgranja24 ,Thanks for your help!
@rcorces ,And I've run this function successfully,Using the function @jgranja24 wrote.
Thanks again for all your help! (I also use R 4.0.1)
liviuspenter commentedon Jul 8, 2020
I can confirm this problem. The fixed function worked for me and I am happy to have found the information in this thread!
My system:
R version 4.0.2 (2020-06-22) -- "Taking Off Again"
Copyright (C) 2020 The R Foundation for Statistical Computing
Platform: x86_64-apple-darwin19.5.0 (64-bit)
rcorces commentedon Jul 8, 2020
@jgranja24 can you make the relevant update(s) in dev? This gets rid of the project summary so if anything downstream uses the summary that will also need to change
ankushs0128 commentedon Aug 7, 2020
Facing the Same Issue on R 4.0.1! Running filterdoublet functions works well
rcorces commentedon Aug 11, 2020
@jgranja24 - can you make this update on
dev?jgranja24 commentedon Oct 15, 2020
This is now fixed on the release branch
devtools::install_github("GreenleafLab/ArchR", ref="release_1.0.0", repos = BiocManager::repositories())Closing this issue, if you have any more troubles please open a new issue!
Best
Jeff